Panaroo
Introduction
Panaroo elaborates the pan-genome and builds presence/absence matrices of samples' annotated genes, starting from Prokka's genome annotation.
For in depth information about Panaroo and its operation, please refer to Panaroo's official guide.
Panaroo's GitHub Page: https://github.com/gtonkinhill/panaroo
Run Analysis Panaroo
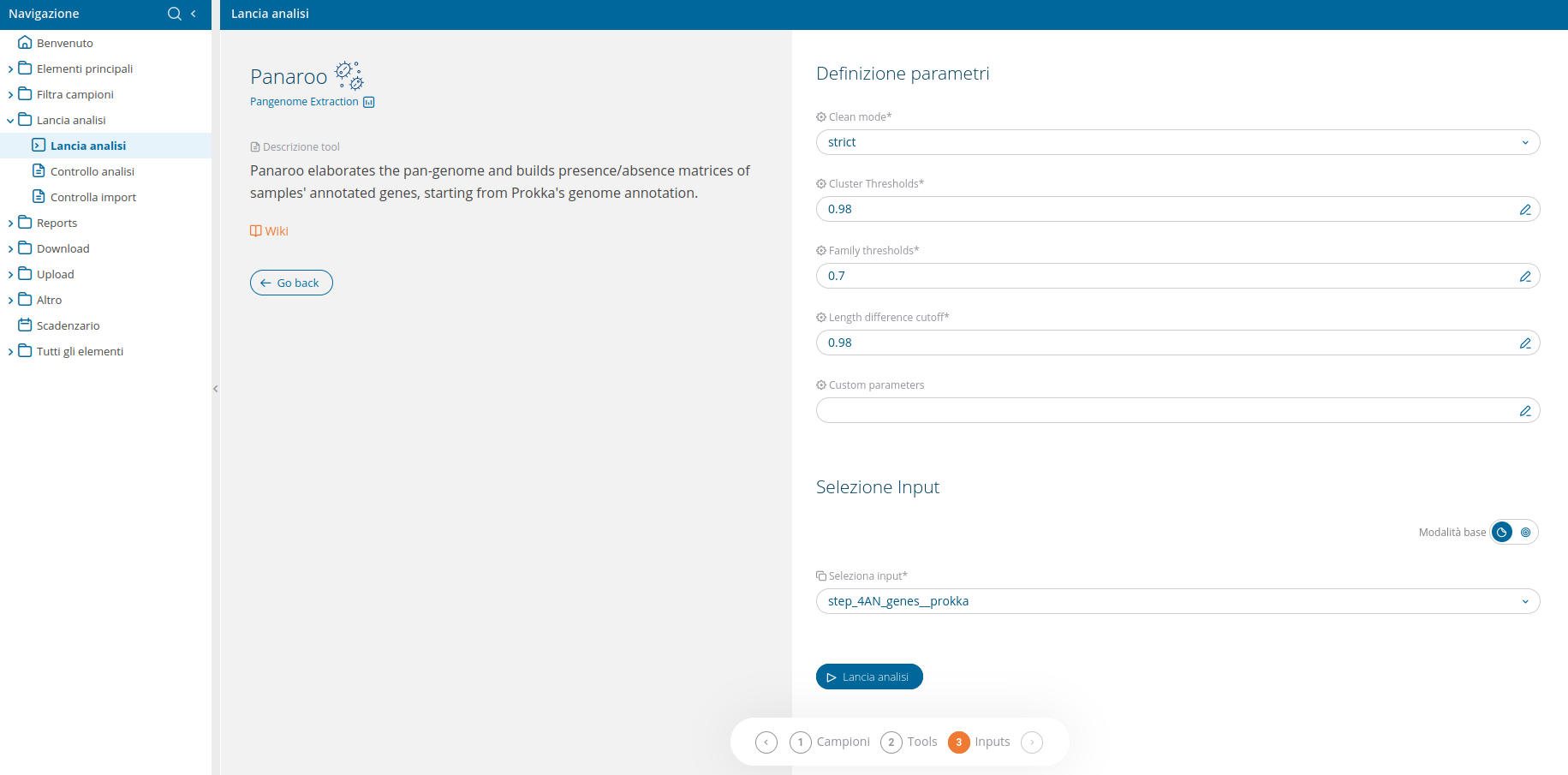
Once the analysis Panaroo has been selected from the run analyses interface, the wizard will present a confirmation UI: there is no need for tool selection, since the only tool available is "Panaroo - An updated pipeline for pangenome investigation".
The input selection wizard will allow to confirm the input for Panaroo, which is deisgned to work on Prokka's output (thus the annotation files from 4AN_genes will be the input). Fields are pre-filled and no further selection by the user is required.
A link to Check analysis will be created after launching the requested analysis. The system will notify the user after a succesful analysis launch and once execution has ended.
Output directory
Please refer to Cohesive's specific Wiki page for information on file download.
The output directory can be reached from the link of the download page or from the link in the analysis summary. The results directory is located directly in the root directory and it contains the following 2 directories:
- meta: ("metadata") contains log and configuration files.
- result: contains the analysis' output files.
The following table lists files created by Panaroo, alongside some useful information. More information on Panaroo's output files are available at the official guide to Panaroo's outputs.
| File | Description | Location |
|---|---|---|
| combined_DNA_CDS.fasta | fasta file of nucleotide sequences from annotated genes and genes identified by Panaroo | results directory |
| combined_protein_CDS.fasta | fasta file of aminoacid sequences from annotated genes and genes identified by Panaroo | results directory |
| combined_protein_GFF3cdhit_out.txt | log of Panaroo's CD-HIT phase | results directory |
| combined_protein_cdhit_out.txt.clstr | CD-HIT cluster info | results directory |
| core_alignment_header.embl | results directory | |
| core_gene_alignment.aln | alignment file | results directory |
| final_graph.gml | pan-genome graph | results directory |
| gene_data.csv | csv table with sequences of annotated genes and corresponding Panaroo internal codes | results directory |
| gene_presence_absence.Rtab | gene presence/absence binary matrix | results directory |
| gene_presence_absence.csv | csv file of gene presence in samples | results directory |
| gene_presence_absence_roary.csv | csv file of gene presence in samples (Roary model) | results directory |
| pan_genome_reference.fa | pan-genome reference fasta for genes in the dataset | results directory |
| pre_filt_graph.gml | raw pan-genome graph | results directory |
| struct_presence_absence.Rtab | presence/absence binary matrix for gene rearrangement events | results directory |
| summary_statistics.txt | metrics summary file | results directory |
Panaroo's authors suggest Cytoscape for graph visualization. more information on pan-genome graph visualization are available at Panaroo's official documentation page.
