1PP_downsampling
Introduction
Downsampling is defined as a process of reduction in read depth (vertical coverage), at specific positions or regions of the genome.
Sequencing protocols can cause stacking of reads for a specific region, leading to excessive data that slows down execution of downstream analyses, while providing no additional information. Downsampling reads prevents extension of calculation times by discarding read pairs until a defined threshold for desired vertical coverage.
Run Analysis 1PP_downsampling
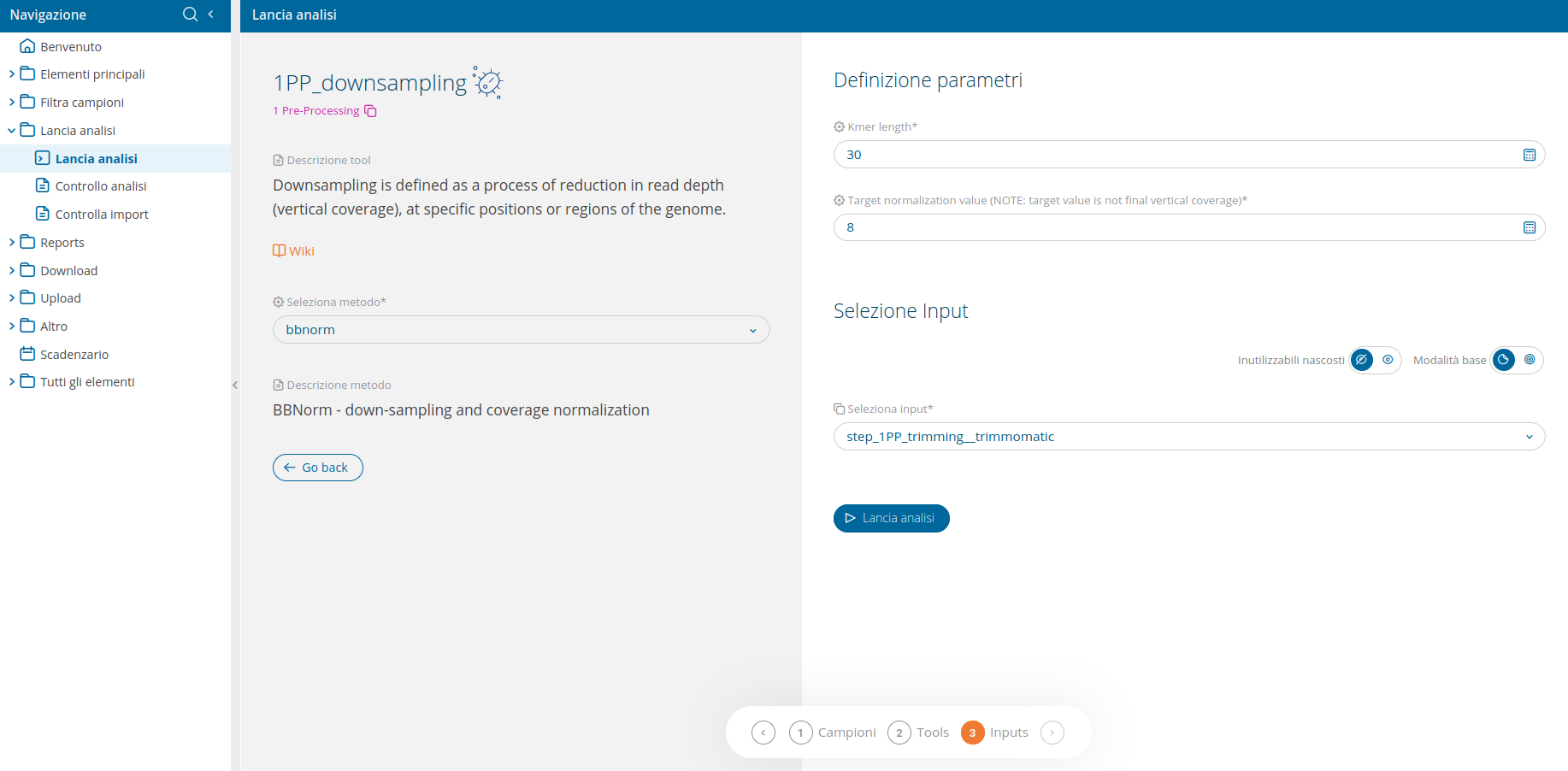
Once the analysis 1PP_downsampling has been selected from the run analyses interface, the user will be able to select which bioinformatic tool to use. The only available tools for this analysis is bbnorm.
The wizard will require input reads and additional parameters to define the target vertical coverage.
The "kmer length" parameter is specific for used samples, while the "Target" parameter is dependent on kmer length and species. As a result, the second parameter has to be determined empirically.
Warning: the second parameter does not correspond to the final verticale coverage.
Two examples for Listeria monocytogenes are listed below.
- species L. monocytogenes, kmer length = 30, target = 31 --> vertical coverage = 40X
- species L. monocytogenes, kmer length = 30, target = 8 --> vertical coverage = 10X
The input selection wizard delivers an advanced input selection mode, to allow selection of all types of supported input files at once.
- step_1PP_trimming
- step_1PP_hostdepl
- step_1PP_downsampling
A link to Check analysis will be created after launching the requested analysis. The system will notify the user after a succesful analysis launch and once execution has ended.
Output directory
Please refer to Cohesive's specific Wiki page for information on file download.
The output directory is available at the link in the download page or at the link presente in the analysis' summary card, and will have the following structure: results > YEAR > ID > 1PP_downsampling > DSXXXXXXXX-DTXXXXXX_bbnorm. At that path there will be 2 directories:
- meta: ("metadata") contains log and configuration files.
- result: contains the analysis' output files.
The table below lists files available in the output directory structure, alongside some useful information.
| File | Description | Location |
|---|---|---|
| DSXXXXXXXX-DTXXXXXX_ID_bbnorm_kXX_tX_R1.fastq.gz | downsampled read 1 (R1) | result directory |
| DSXXXXXXXX-DTXXXXXX_ID_bbnorm_kXX_tX_R2.fastq.gz | downsampled read 2 (R2) | result directory |
