Depletion & de novo Pipeline
Introduction
The Depletion & de novo pipeline performs consecutively host depletion (1PP_hostdepl) and de novo assembly (2AS_denovo).
Run Depletion & de novo Pipeline
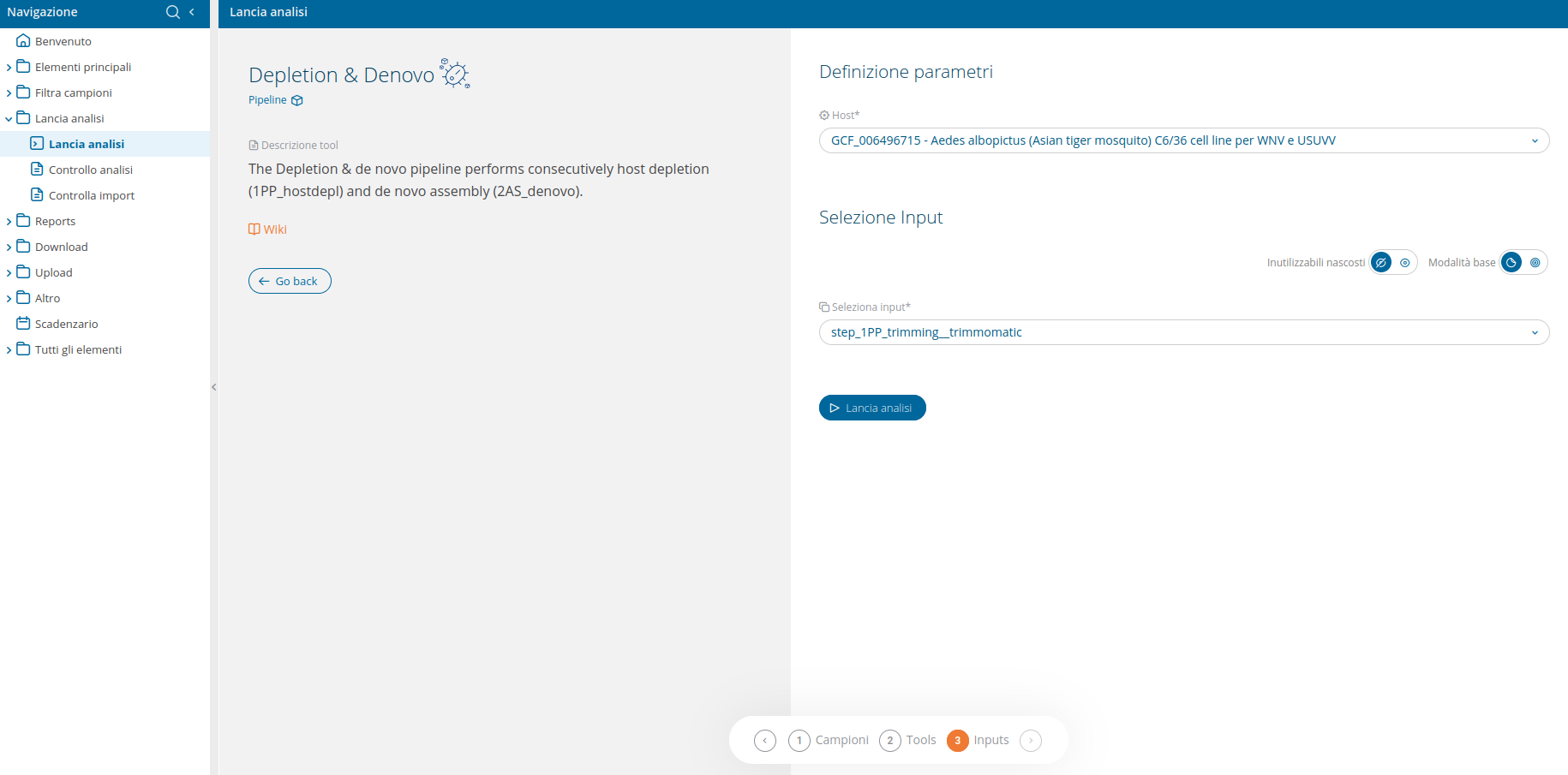
The filter at the top of the run analyses page allows to display only the pipelines. Once the Depletion & de novo pipeline has been selected, a confirmation interface will be displayed.
The pipeline performs the following 2 analyses:
- Depletion of host sequences with bowtie (1PP_hostdepl__bowtie);
- de novo assembly with Spades (2AS_denovo__spades) e controllo qualità con Quast.
Inputs are the same as those available for host depletion, i.e. the host's reference genome and the fastq reads:
The input selection UI delivers an advanced input selection mode, to allow selection of all types of supported input files at once.
A link to Check analysis will be created after launching the requested pipeline. The system will notify the user after a succesful pipeline launch and once execution has ended.
Results
Output files from the "Deplezione & de novo" pipeline are the same as those produced by the single analyses included in it, in the same directory structure:
