Filtering & de novo
Introduction
The Filtering & de novo pipeline removes reads that do not map on the selected reference (1PP_filtering) and performs de novo assembly (2AS_denovo).
Run Filtering & de novo Pipeline
The filter at the top of the run analyses page allows to display only the pipelines. Once the Filtering & de novo pipeline has been selected, a confirmation interface will be displayed.
Filtering & de novo performs:
- removal of reads that do not map to the reference with Bowtie (1PP_filtering__bowtie)
- de novo assembly with SPAdes (2AS_denovo__spades)
Accepted inputs can be from pre-processing analyses:
- depleted reads (step_1PP_hostdepl)
- downsampled reads (step_1PP_downsampling)
- trimmed reads (step_1PP_trimming)
- filtered reads (step_1PP_filtering)
The input selection UI delivers an advanced input selection mode, to allow selection of all types of supported input files at once.
The pipeline also requires to specify, through the dedicated wizard, a reference genome to use for the first analysis of the pipeline: filtering (1PP_filtering).
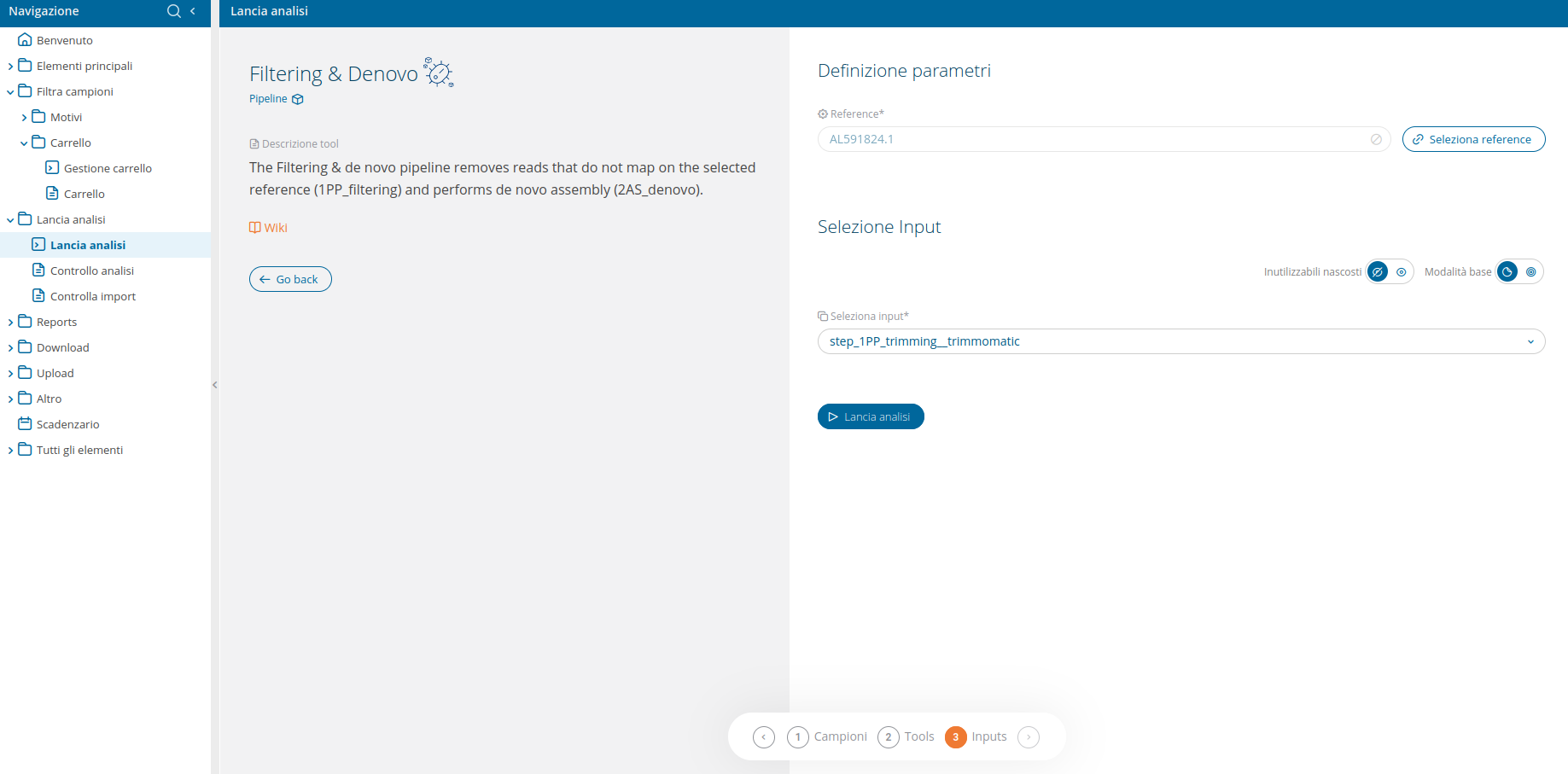
A link to Check analysis will be created after launching the requested pipeline. The system will notify the user after a succesful pipeline launch and once execution has ended.
Results
Output files from the "Filtering & de novo" pipeline are the same as those produced by the single analyses included in it, in the same directory structure:
