1PP_trimming
Introduction
The trimming step removes low quality nucleotide residues from reads produced by the sequencer. The 1PP_trimming analysis in Cohesive includes the execution of trimmomatic and fastqc. The latter gives quality metrics about the reads, i.e. raw reads (before trimming) and produced trimmed reads.
Run Analysis 1PP_trimming
Once the analysis 1PP_trimming has been selected from the run analyses interface, the user will be able to select which bioinformatic tool to use. The available tool for 1PP_trimming is:
- trimmomatic - Read trimming tool for Illumina NGS data
Input is selected in the wizard's last section: "step_0SQ_rawreads__fastq" is for internal fastq files from sequencers, (code 20XX.TE.XXXX.X.X), while "step_0SQ_rawreads__external", is for imported fastq files (code 20XX.EXT.XXXX.X.X). The input selection UI also delivers an advanced input selection mode, to allow selection of all types of supported input files at once.
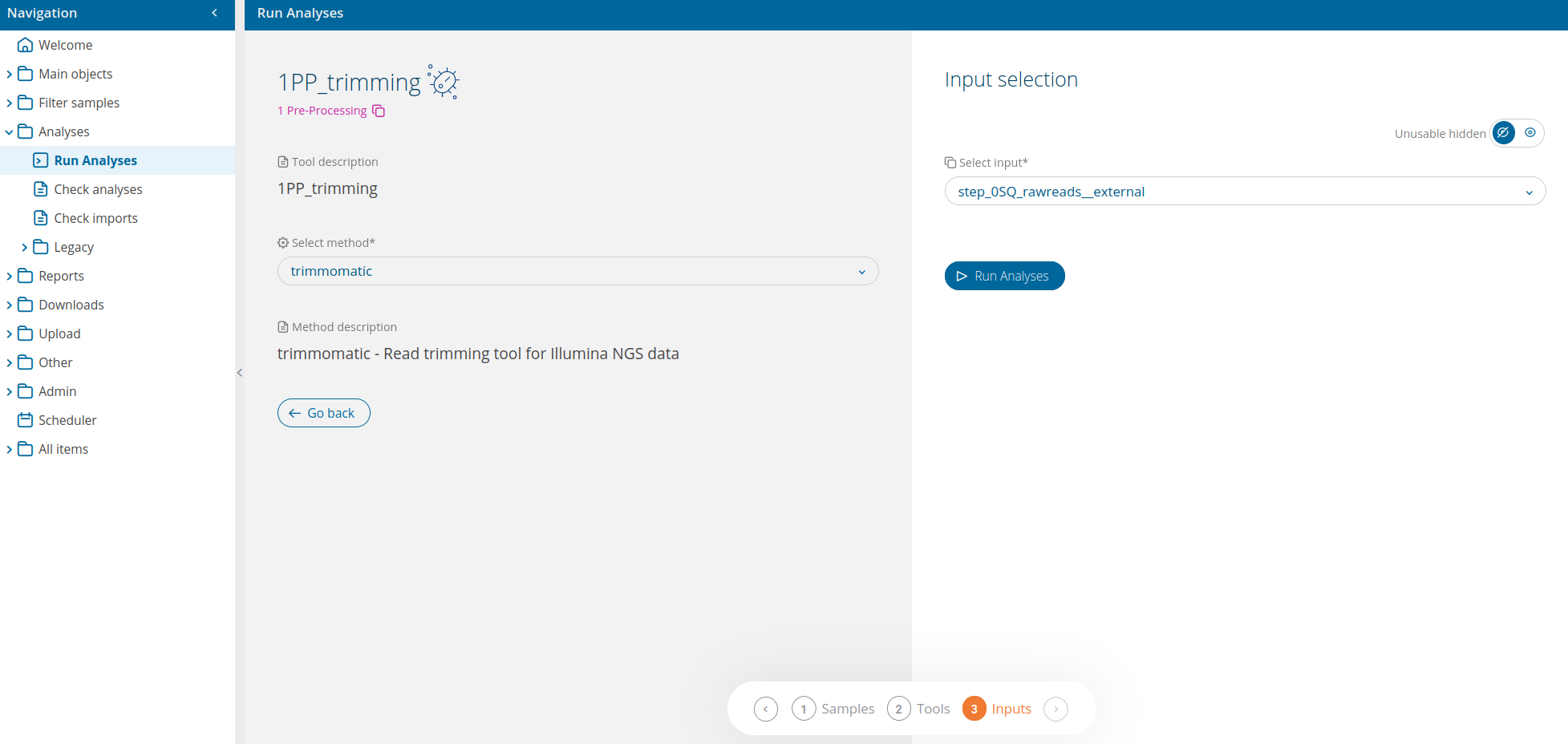
A link to Check analysis will be created after launching the requested analysis. The system will notify the user after a succesful analysis launch and once execution has ended.
Output directory
Please refer to Cohesive's specific Wiki page for information on file download.
The output directory is available at the link in the download page or at the link presente in the analysis' summary card, and will have the following structure: results > YEAR > ID > 1PP_trimming > DSXXXXXXXX-DTXXXXXX_trimmomatic.
At that path there will be 3 directories:
- meta: ("metadata") contains log and configuration files.
- qc: ("quality check") it contains 2 directories (meta and result). In this case quality check is performed with fastqc.
- result: contains the analysis' output files.
The table below lists files available in the output directory structure, alongside some useful information.
| File | Description | Location |
|---|---|---|
| DSXXXXXXXX-DTXXXXXX_ID_R1_trimmomatic.fastq | trimmed read 1 (R1) | result directory |
| DSXXXXXXXX-DTXXXXXX_ID_R2_trimmomatic.fastq | trimmed read 2 (R2) | result directory |
| DSXXXXXXXX-DTXXXXXX_ID_unpaired_trimmomatic.fastq | trimmed unpaired reads | result directory |
| DSXXXXXXXX-DTXXXXXX_ID_R1_trimmomatic_fastqc.html | reads R1 quality | qc directory > result |
| DSXXXXXXXX-DTXXXXXX_ID_R1_trimmomatic_fastqc.zip | quality R1 (zip file) | qc directory > result |
| DSXXXXXXXX-DTXXXXXX_ID_R2_trimmomatic_fastqc.html | reads R2 quality | qc directory > result |
| DSXXXXXXXX-DTXXXXXX_ID_R2_trimmomatic_fastqc.zip | quality R2 (zip file) | qc directory > result |
| DSXXXXXXXX-DTXXXXXX_ID_unpaired_trimmomatic_fastqc.html | unpaired reads quality | qc directory > result |
| DSXXXXXXXX-DTXXXXXX_ID_unpaired_trimmomatic_fastqc.zip | unpaired reads (zip file) quality | qc directory > result |
